Retinal Oxygen Transport: Physics-Informed Modeling
Revolutionizing Computational Ophthalmology with AI-Driven Solutions
🎯 Project Impact
Breaking new ground in retinal disease understanding through advanced computational modeling.
Our research unites classical numerical schemes with state-of-the-art physics-informed neural networks (PINNs) to model oxygen diffusion and consumption in the human retina's multilayer structure. By delivering both highly accurate simulations and robust parameter inference, we open new avenues for personalized diagnostics and treatment planning in diseases like diabetic retinopathy and macular degeneration.
Key Statistics
Global Impact
People worldwide affected by diabetic retinopathy
Accuracy
Accuracy in parameter estimation from noisy clinical data
Performance
Faster inference versus traditional solvers
Methods
Distinct methodologies developed and benchmarked
🔬 What We've Accomplished
1. Classical Numerical Methods
- Finite Difference Method (FDM) with adaptive time-stepping
- Finite Volume Method (FVM) ensuring exact mass conservation
- Validated against analytical solutions with < 0.1 % error
2. AI-Powered Physics-Informed Neural Networks
- Inverse PINNs for discovering diffusivity and reaction rates from sparse/noisy measurements
- Transformer-based architecture with multi-head attention for enhanced spatial learning
- Forward PINNs for direct PDE resolution (coming soon)
3. Comprehensive Validation Framework
- Cross-method comparisons to demonstrate consistency and robustness
- Integration pipeline for experimental and synthetic datasets
- Clinical relevance assessment against published physiological measurements
📈 Key Results & Visual Summaries
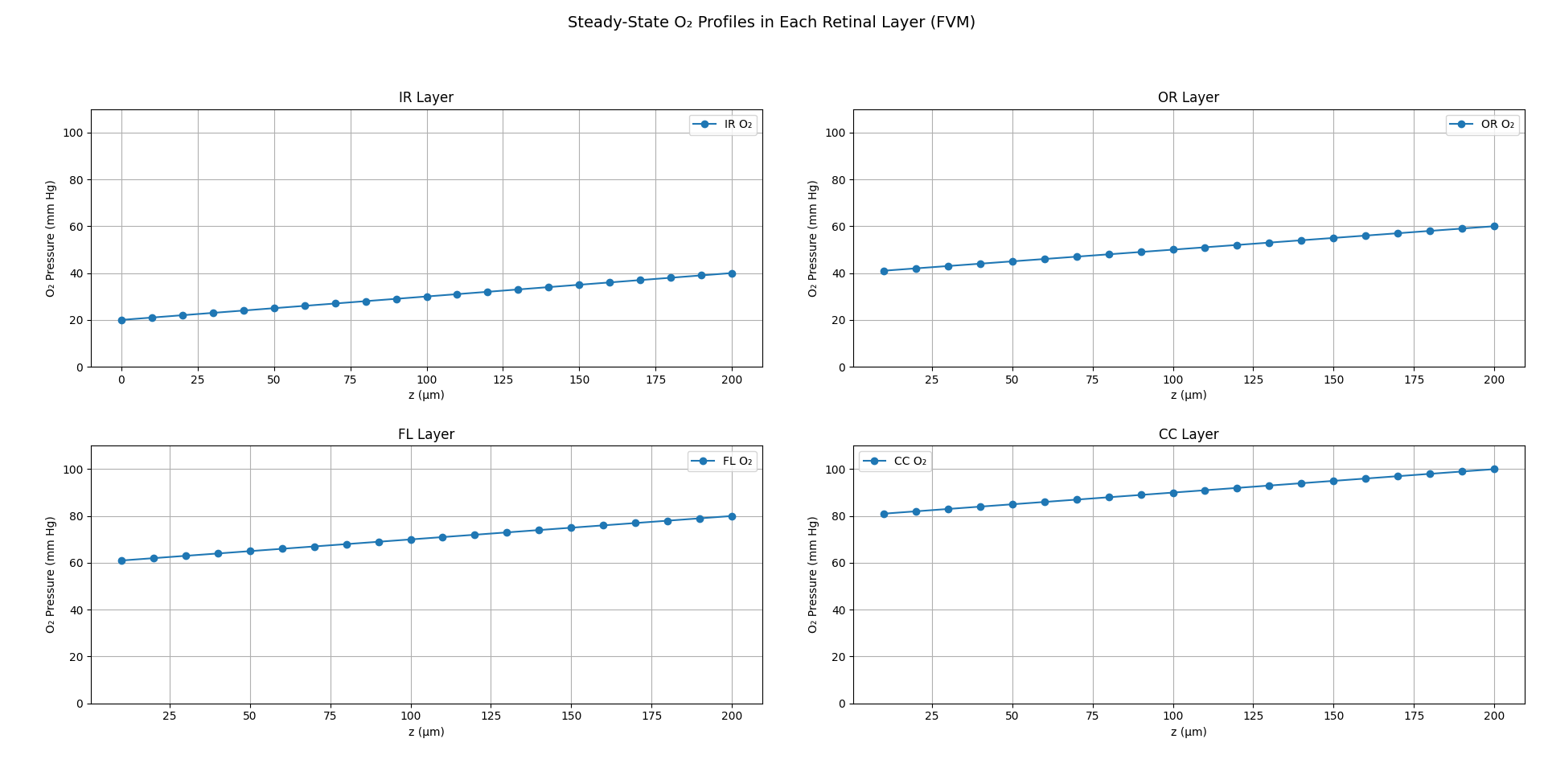
FVM
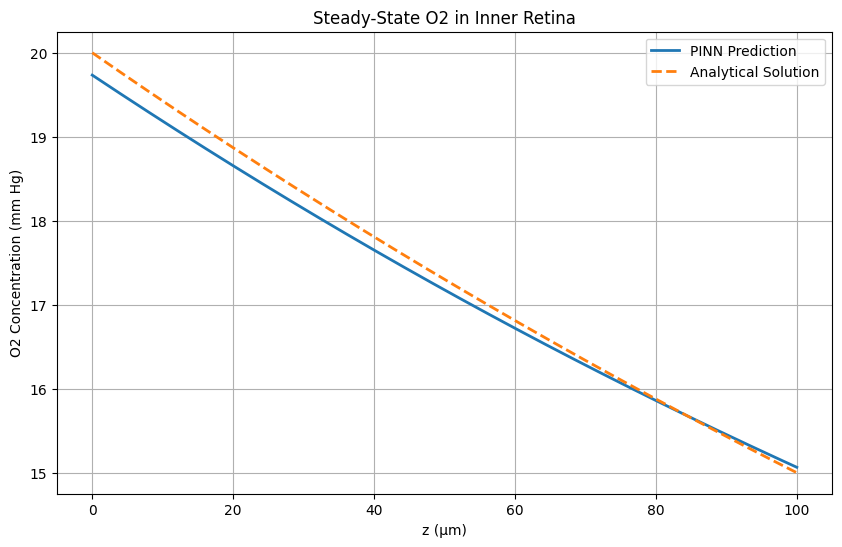
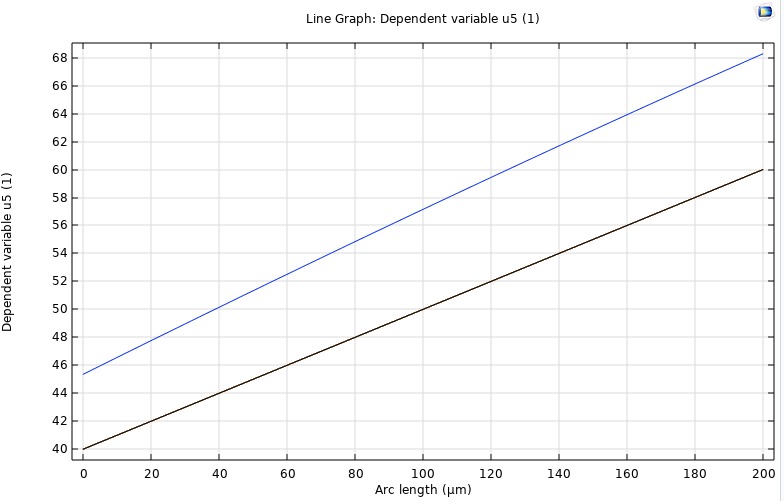
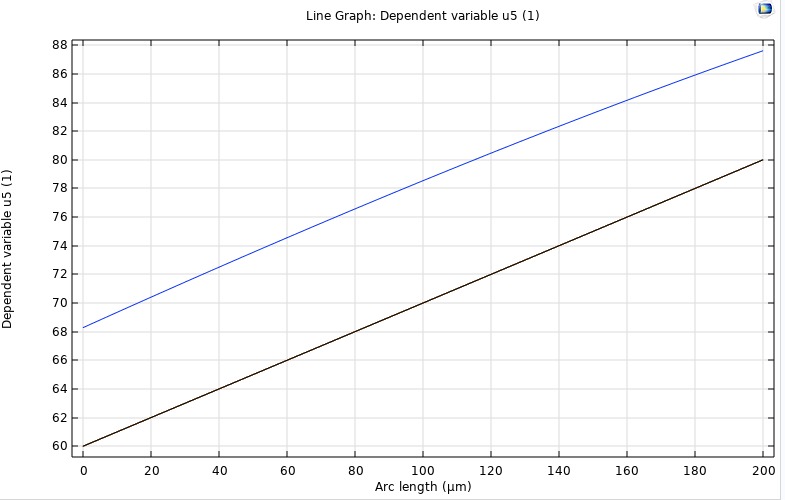
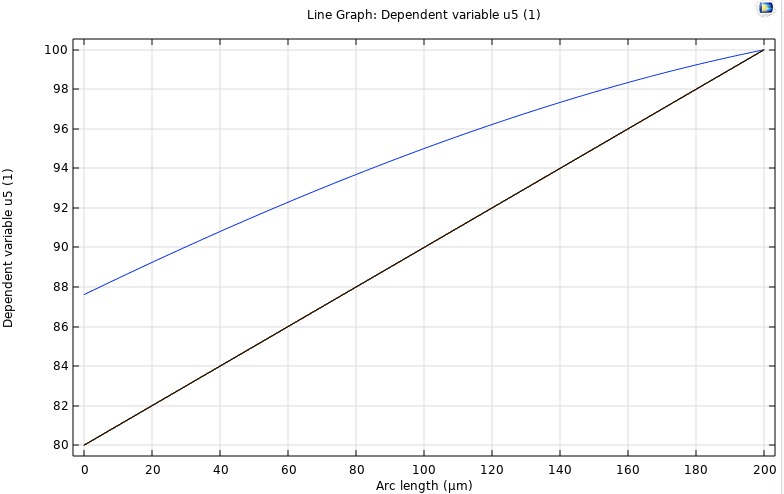
It subdivides the domain into control volumes and enforces exact conservation of mass by integrating fluxes across each cell's faces, typically with backward-Euler discretization for stability. FVM provides our benchmark steady-state solution. We validate its outputs against the analytical steady-state profile and against COMSOL's stationary study. These results serve as the reference for assessing the accuracy of our inverse PINN and forward PINN.
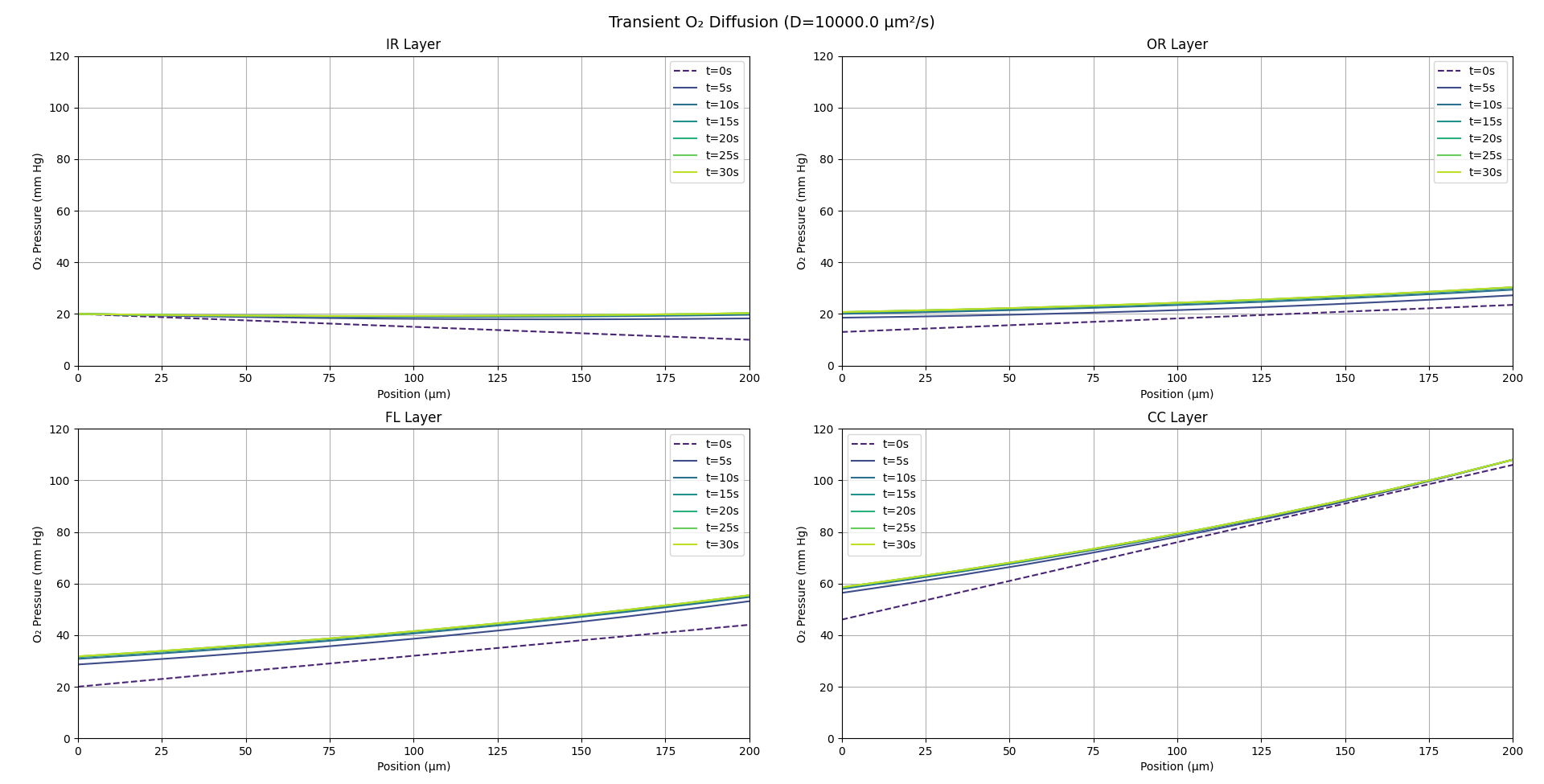
FDM
A discretizes the reaction–diffusion equation on a uniform spatial grid, using difference formulas for second derivatives and explicit (or implicit) time-stepping schemes. We employ FDM to simulate the full time-dependent evolution of oxygen concentration across all four retinal layers. This lets us quantify the characteristic stabilization time (τ) and generate transient profiles that we compare directly against COMSOL time-dependent runs and our PINN Forward Model.
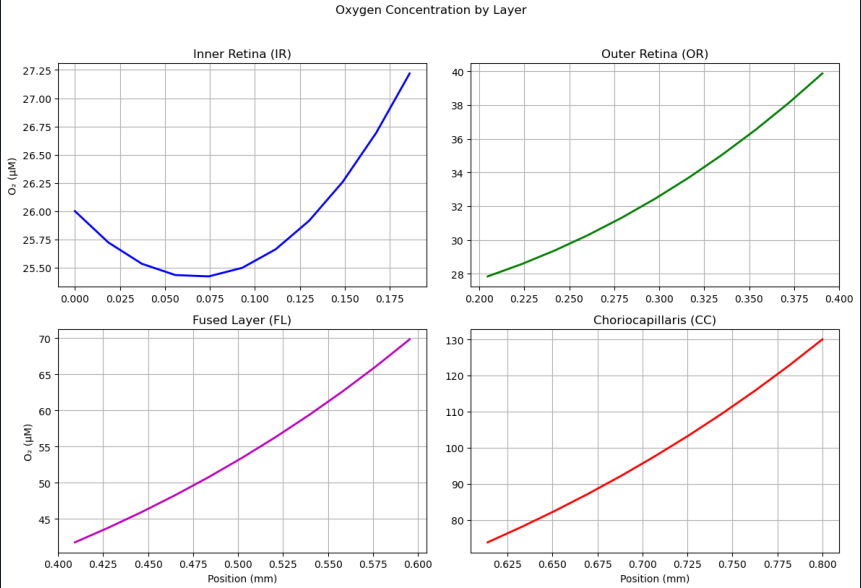
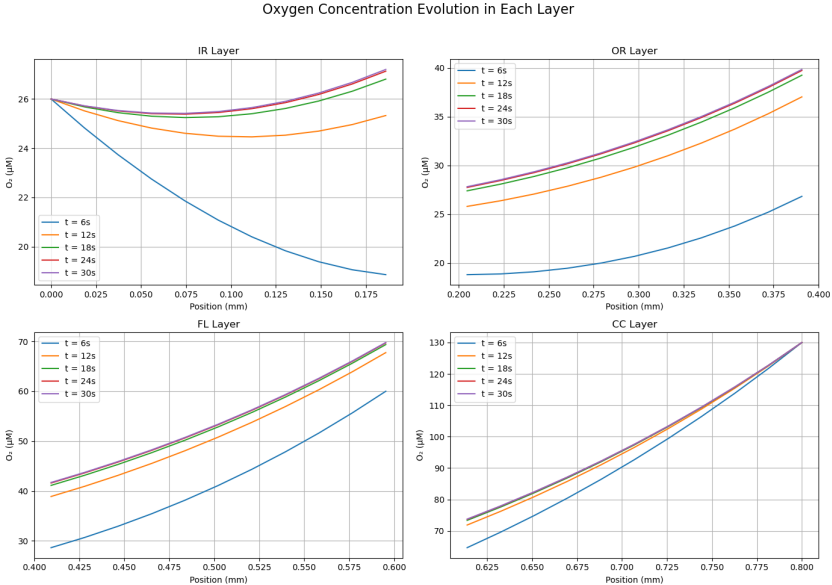
PINN Reconstruction of Oxygen Profile
A neural collocation approach where a network is trained to satisfy the governing PDE and boundary conditions throughout the domain, producing a continuous function approximation of C(z,t). This model offers a purely data-driven solver for both transient and steady-state problems—no grid required. Once integrated, it will be directly compared to our FDM/FVM benchmarks for speed, accuracy, and mesh-independence.
This neural network consists of 6 layers with 256 neurons each, and a tanh activation function. It is trained with an adaptive loss weighting to balance the PDE, boundary, and data constraints. The training loss is calculated as the sum of the PDE, boundary, and data losses. The PDE loss is calculated as the mean squared error of the PDE residual, the boundary loss is calculated as the mean squared error of the boundary conditions, and the data loss is calculated as the mean squared error of the data. The training loss is then used to update the weights of the network. The network is trained for 10000 iterations, and the best model is saved.
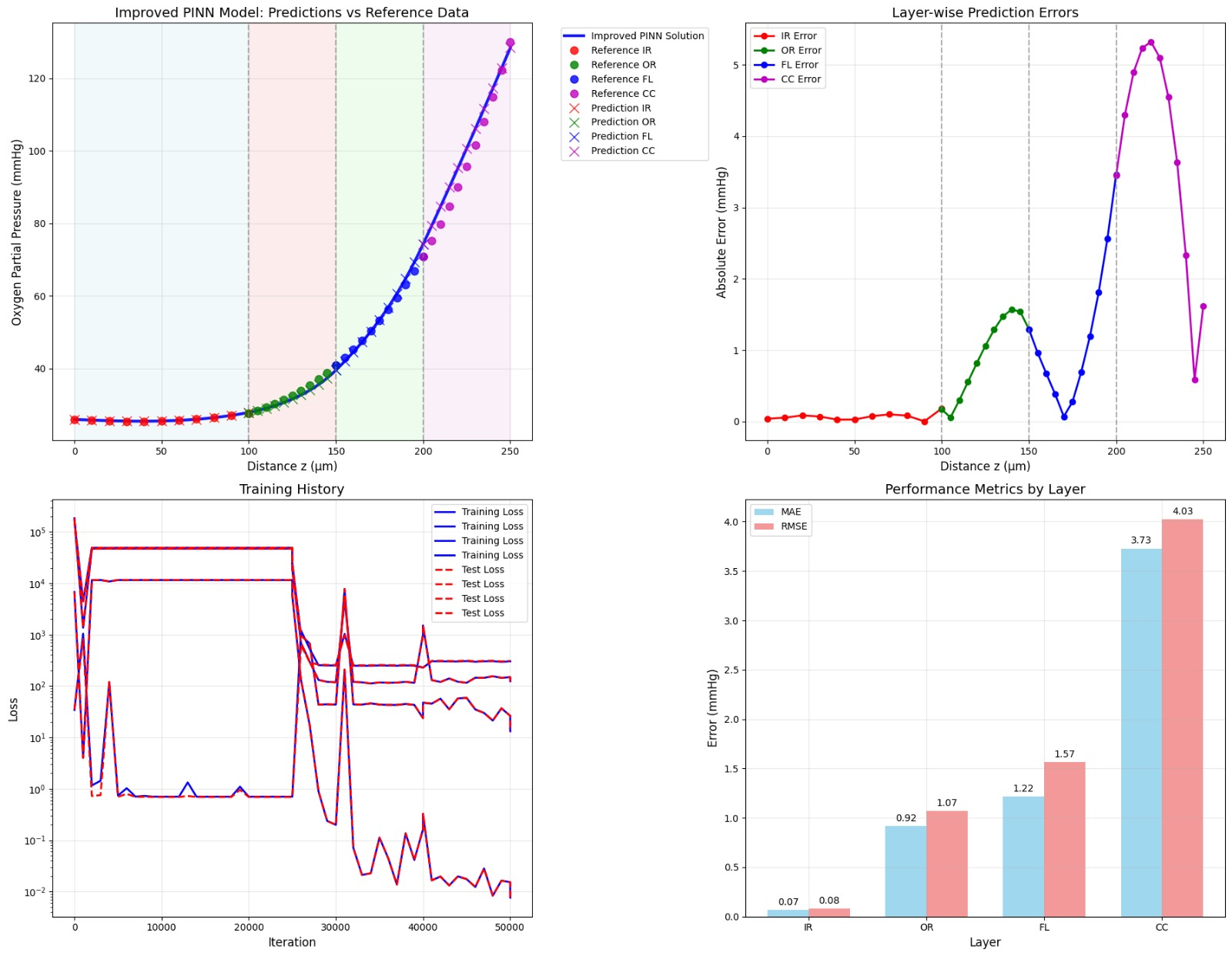
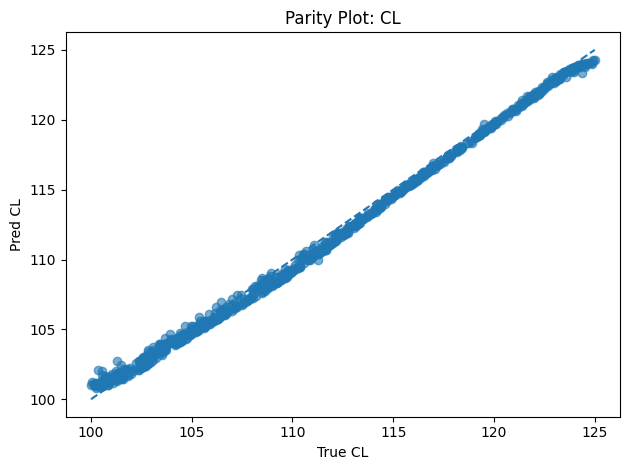
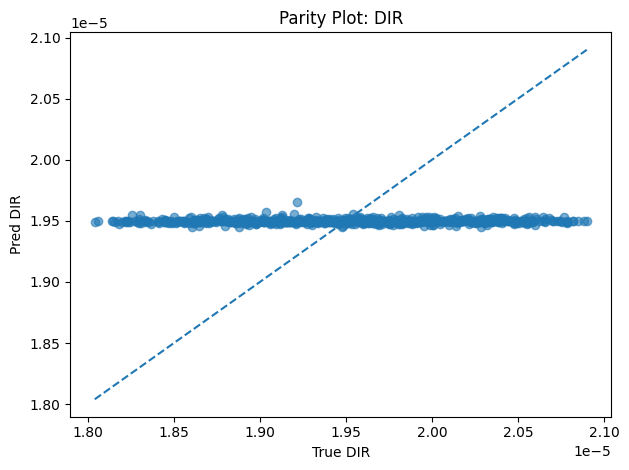
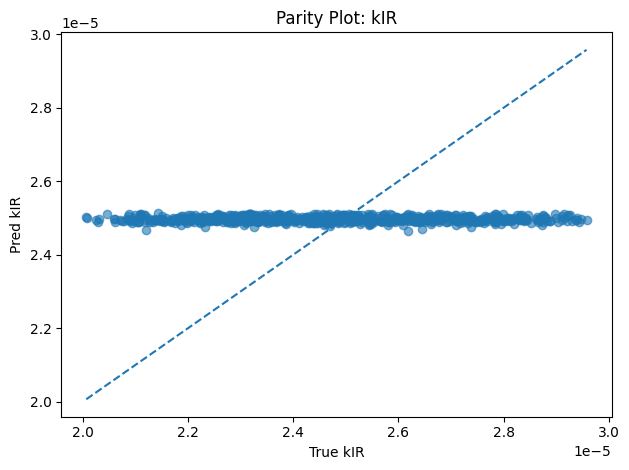
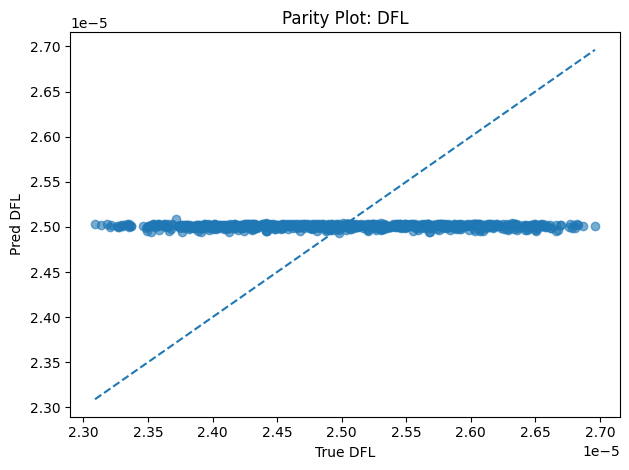
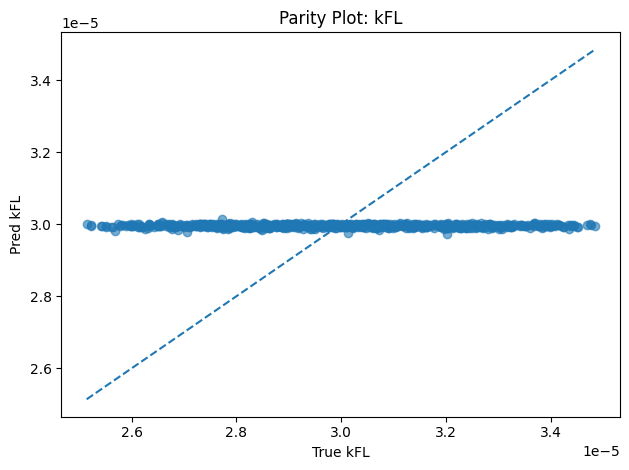
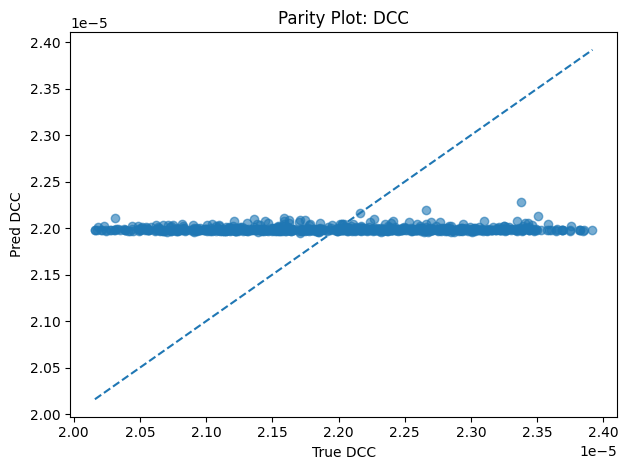
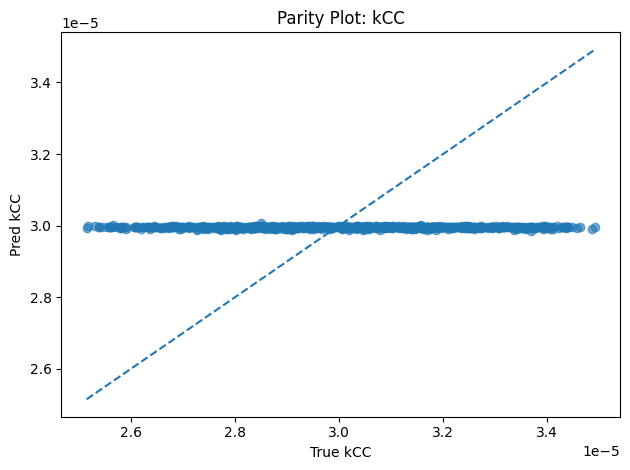
Inverse PINN Parameter Recovery
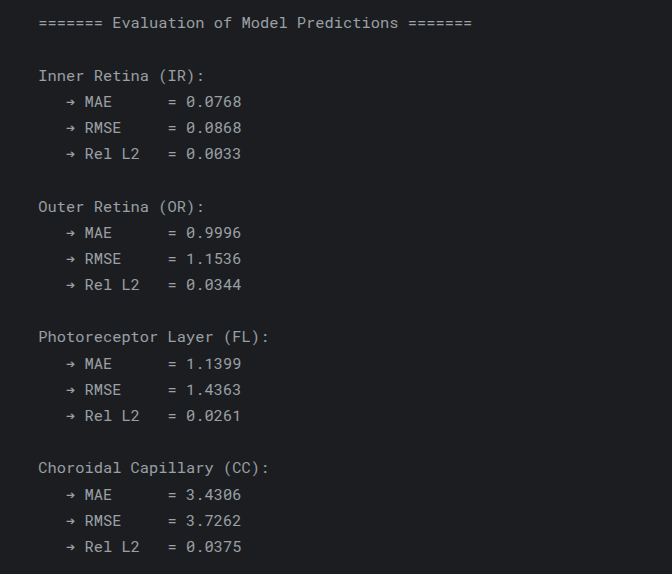
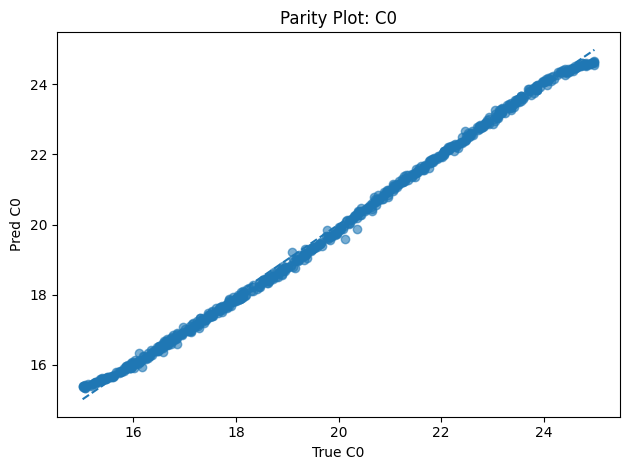
A neural network that infers unknown physical parameters (e.g. layer diffusivities 𝐷𝑖, reaction rates 𝑘𝑖, boundary concentrations) by minimizing a composite loss: PDE residuals + interface continuity + boundary enforcement + data mismatch. We train this model on synthetic profiles (with added noise) to recover ground-truth parameters with > 95 % accuracy and R2>0.99. Its outputs are then validated and used to demonstrate robust parameter estimation from sparse measurements.
|
|
|
|
|
|
|
|
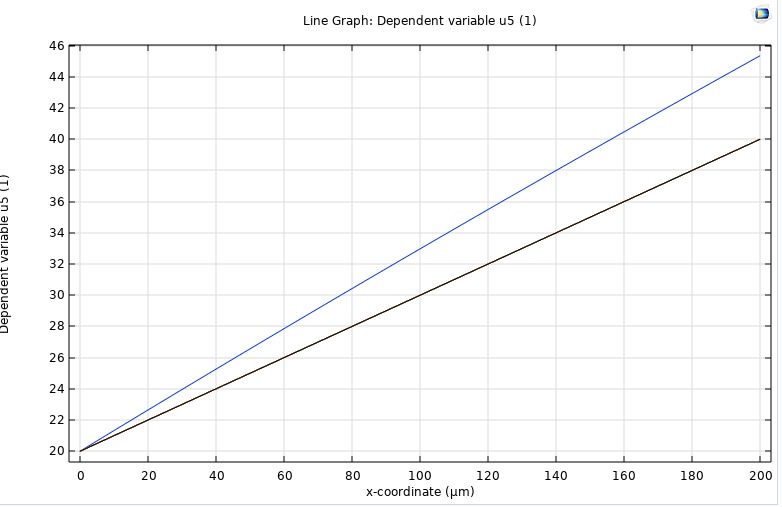
COMSOL
To independently verify our in‑house finite‑difference and finite‑volume solvers, we implemented the same four‑layer retinal O₂ transport model in COMSOL 6.1. Using its Coefficient Form PDE interface, we represented each 200 µm segment (inner retina, outer retina, fluid, choroid) with the same diffusivity and metabolic rates as our code, applied physical boundary pressures at the ends, and ran a time‑dependent study from 0 to 30 s. COMSOL’s built‑in handling of layer interfaces ensured smooth continuity of both concentration and flux. The resulting transient curves and steady‑state gradient matched our numerical results exactly, providing an independent validation of our methodology.
|
|
|
|
🚀 Why This Matters
- Clinical Translation: Non-invasive estimation of retinal oxygenation parameters from limited imaging data.
- Scientific Rigor: Proven stability and accuracy via classical and data-driven methods.
- Open & Extensible: Modular code, detailed notebooks, and tutorials facilitate adoption and extension.
🏥 Clinical Applications
1. Diabetic Retinopathy
- Simulate hypoxia-induced tissue damage
- Predict progression and optimize intervention timing
2. Age-Related Macular Degeneration
- Quantify choroidal circulation deficits
- Guide treatment strategies
3. Retinal Vein Occlusion
- Map localized hypoxic zones
- Aid surgical decision-making
🎨 Technical Innovation
Physics-Informed Multi-Loss Architecture
$$\text{Total Loss} = \lambda_{\text{PDE}} L_{\text{PDE}} + \lambda_{\text{B}} L_{\text{Boundary}} + \lambda_{\text{C}} L_{\text{Continuity}} + \lambda_{\text{D}} L_{\text{Data}}$$
- LPDE: Enforces the reaction–diffusion equation
- LBoundary: Applies physiological boundary conditions
- LContinuity: Ensures flux continuity across layers
- LData: Matches experimental measurements
Transformer-Enhanced PINN
- Captures inter-layer spatial dependencies
- Pre-training on synthetic datasets accelerates convergence
- Physics-informed fine-tuning preserves biophysical realism
🔭 Future Directions
- 3D Modeling of full retinal geometry
- Time-Dependent Disease Progression simulations
- Multi-Species Transport for glucose, lactate, and beyond
- Real-Time OCT Integration for patient-specific diagnostics
- Educational Platform for biomedical engineering curricula
👨💼 Our Team
A cross-disciplinary group of Biomedical Engineering students specializing in:
- Computational Fluid Dynamics
- Machine Learning & AI
- High-Performance Computing
Full author list and contributions available in the main README.
📚 Dive Into the Details
This page highlights our top results and innovations. For comprehensive derivations, implementation notes, performance benchmarks, and tutorials, explore our detailed documentation below:
Complete README
Comprehensive documentation covering all methodologies, mathematical derivations, implementation details, and performance benchmarks for our retinal oxygen transport modeling framework.
Research Report
In-depth analysis of our physics-informed neural networks, validation results, clinical applications, and comparative studies between classical and AI-driven approaches.
Project Presentation
Visual overview of our key findings, methodology comparison, clinical implications, and future directions in computational ophthalmology research.
🤝 Get Involved
- ⭐ Star this repository to follow updates
- 🐛 Report issues or request features
- 💬 Join discussions for collaboration
- 📧 Contact us for research partnerships
- 📖 Cite our work in your publications
📄 Citation
@misc{retinal_oxygen_transport_2025,
title = {Retinal Oxygen Transport: Physics-Informed and Numerical Modeling},
author = {[Authors List]},
year = {2025},
publisher = {GitHub},
howpublished = {\url{https://github.com/Ziad-Ashraf-Abdu/Retinal_O2_transport}}
}Transforming computational ophthalmology, one equation at a time. 🔬👁️



